Académique Documents
Professionnel Documents
Culture Documents
15 Trimerization
Transféré par
api-37060270 évaluation0% ont trouvé ce document utile (0 vote)
18 vues8 pagesGilmore R., Coffey M.C., Leone G., McLure K., Lee P.W.K. (1996) Cotranslational trimerization of the reovirus cell attachment protein. EMBO J. 15, 2651 – 2658.
Titre original
15-Trimerization
Copyright
© Attribution Non-Commercial (BY-NC)
Formats disponibles
PDF ou lisez en ligne sur Scribd
Partager ce document
Partager ou intégrer le document
Avez-vous trouvé ce document utile ?
Ce contenu est-il inapproprié ?
Signaler ce documentGilmore R., Coffey M.C., Leone G., McLure K., Lee P.W.K. (1996) Cotranslational trimerization of the reovirus cell attachment protein. EMBO J. 15, 2651 – 2658.
Droits d'auteur :
Attribution Non-Commercial (BY-NC)
Formats disponibles
Téléchargez comme PDF ou lisez en ligne sur Scribd
0 évaluation0% ont trouvé ce document utile (0 vote)
18 vues8 pages15 Trimerization
Transféré par
api-3706027Gilmore R., Coffey M.C., Leone G., McLure K., Lee P.W.K. (1996) Cotranslational trimerization of the reovirus cell attachment protein. EMBO J. 15, 2651 – 2658.
Droits d'auteur :
Attribution Non-Commercial (BY-NC)
Formats disponibles
Téléchargez comme PDF ou lisez en ligne sur Scribd
Vous êtes sur la page 1sur 8
‘The EMBO Journal vol.18 10.11 pp.2651-2658, 1998
Co-translational trimerization of the reovirus cell
attachment protein
Ross Gilmore, Matthew C.Coffey,
Gustavo Leone, Kevin McLure and
Patrick W.K.Lee’
Depurtment of Microbiology snd Infectious Diseases, Univesity of
Calgary Health Sciences Gente, Calgary, Albert, Canala TON NI
‘Conespomding author
The reovirus cell attachment protein, ol, is a trimer
with a ‘lollipop’ structure. Recent findings indicate
that the N-terminal fibrous tail and the C-terminal
globular head each possess a distinct trimerization
domain, The region responsible for N-terminal
trimerization (formation of a triple a-helical coiled-
coil) is located at the N-terminal one-third of ol. In
this study, we investigated the temporality and ATP.
requirement of this trimerization event in the context,
of 61 biogenesis. Jn vitro co-synthesis of the full-length,
(FL) and a C-terminally truncated (444) o1 protein,
revealed a preference for homotrimer over hetero
trimer formation, suggesting that assembly at the
N-terminus occurs co-translationally. This was cor-
roborated by the observation that polysome-associated
Gl chains were trimeric as well as monomeric. Trun-
cated proteins (d234 and d294) with C-terminal dele-
tions exceeding half the length of o1 were found to
trimerize post-translationally. This trimerization did
not require ATP since it proceeded normally in the
presence of apyrase. In contrast, formation of stable
FL of trimers was inhibited by apyrase treatment.
Collectively, our data suggest that assembly of nascent
Ol chains at the N-terminus is intrinsically ATP inde-
pendent, and occurs co-translationally when the ribo-
somes have traversed past the midpoint of the mRNA.
Kewords: protein oligomerization/rabbit reticulocyte
Jysate/reovirus protein G1
Introdu
Significant advances have been made in recent years in
the search for factors and parameters that dictate the
correct folding of polypeptide chains. While the general
concept that the final conformation of a protein is deter-
mined by its amino acid sequence still holds true (Anfinsen,
1973), there is now overwhelming evidence that, in the
call, protein folding is mediated by molecular chaperone
proteins (reviewed by Georgopoulos and Welch, 1993;
Hendrick and Hartl, 1993; Ellis, 1994; Hartl et al
1994). These chaperones assist protein folding in an ATP-
dependent manner by preventing and reversing. hydro-
phobic interactions that lead to aggregates or misfolded
conformations. The initial demonstration by Beckmann
1a. (1990) that nascent polypeptides in the mammalian
© Oxford Univorsity Press
eytosol associate with Hsp70 has led to the hypothesis,
that protein folding begins during biogenesis. Using firefly
luciferase synthesized in rabbit reticulocyte lysate as a
model system, Frydman et al. (1994) found that the
growing polypeptide interacts, in an orderly manner, with
a specific set of molecular chaperones, including Hsp70,
Hsp40 and the chaperonin TCP-1 (t-complex polypeptide
1) complex (TRC), Their data suggest that Hsp70, in
cooperation with Hsp40, is able to bind to very short
nascent chains (~40 exposed residues), whereas TRIC
interacts only with longer chains (~100-150 residues).
Co-translational folding occurs once the nascent chain has,
attained a length of ~250-300 residues, but the rest of the
550 residue long protein undergoes maturation folding only
after release from the ribosome. That nascent luciferase
peptides start to attain their native tertiary structure during
protein synthesis on the ribosomes was also deduced
independently by Kolb ef al, (1994) who carried out
continuous monitoring of the enzymatic activity of newly
synthesized luciferase in a wheat germ cell-free translation
system. Using an Escherichia coli eell-rce system,
Fedorov and Balwin (1995) have shown that the B-subunit
of bacterial luciferase also undergoes co-translational fold
ing, which presumably accounts for the rapid formation
of the native structure in the cell. The demonstration that
heme binds to nascent globin chains suggests that globin
also folds eo-translationally (Komar et al., 1993).
While the biogenetic pathway of monomeric proteins
is being revealed in terms of co- and post-translational
folding and of the involvement of chaperones in these
processes, surprisingly litle is known about such processes
for homooligomerie proteins in the eytosol. In this regard, a
most fundamental question hitherto unanswered is whether
these proteins assemble co-translationally or post-transla
tionally. In the case of membrane-bound homooligomers,
such as the influenza virus hemagglutinin and the vesicular
stomatitis virus (VSV) G protein, available evidence
suggests that newly synthesized full-length proteins are
anchored to the endoplasmic reticulum (ER) as partially
folded monomers which subsequently undergo assembly
with concomitant maturation folding (reviewed by Doms
et al., 1993). This post-translational assembly mechanism
is unlikely to be adopted by cytosolic oligomers in view
of the lack of any confining structure equivalent to the
ER, For these proteins, co-translational assembly with the
initial interaction occurring between nascent chains would
represent a logical, if not the sole, altemative. In the
present study, we test this hypothesis by examining. the
initial assembly step that Teads to the generation of the
trimerie reovirus cell attachment protein (protein 1),
The reovirus 61 trimer is located at the 12 vertices of
the icosahedral virion (Lee et ai., 1981; Furlong et al,
1988). It is highly asymmetric with an N-terminal fibrous
tail thatis anchored to the virion, and a C-terminal globular
2651
RGilmore et al
pulse _chase
A BAB
— — Itrimer
= — | monomer
123 4
Fig. 1, Pulse-chase analysis of full-length a (A) and a C-terminal
tmuneated 1 (444) (B) synthesized in vio, Transcripts encoding these
‘wo proteins were. translated separately in rabbit reticulocyte lysate in
the presence of "S]methionine for 7 min (pulse) and then subjected
to ultacentrifupation at 4°C to pellet the polysomes. The superatants
(pulse), which contained no translation activity, were then incuaed at
37°C for 60) min (chase). Both pulse and chase samples were analyzed
by SDS-PAGE under now-dssecating conditions which allowed for
te detection of timerie 01 forms stabilized via the N-terminss but
ot the C-terminus (i. preincubation of samples in peotein sample
butferat 97°C, followed by electrophoresis caried out at °C),
‘head that interacts with the cell receptor (Banerjea et al,
1988; Furlong et al., 1988; Yeung er al. 1989; Fraser
et al, 1990; Mah et a, 1990; Duncan et al, 1991; Leone
eal, 199 lab; Strong et al, 1991). Evidence from
in vitro translation studies has revealed that these two
structurally distinct domains are generated by independent
trimerization events (Leone et al, 1992), The two domains
also differ in stability, with the N-terminal trimeric
Gchelical coiled-coil being significantly more stable than
the C-terminal trimeric globular head (Strong es al., 1991;
Tumer et al., 1992), These characteristies, together with
the relative ease with which inv vitro-generated 61 folding
and oligomerization intermediates can be resolved and
identified using SDS-PAGE under non-dissociating condi-
tions (Leone e7 al., 1992), have made Gl an attractive
model system for the study of the sequence of events
involved in the folding and assembly of oligomeric proteins
in the cytosol. In this study, we examined the initial
(N-terminal) assembly step of the 61 trimer in terms of
‘temporality (co-translational versus post-translational) and
ATP requirement by analyzing full-length and various
truncated G1 forms synthesized in rabbit reticulocyte
lysate. Ourresulls suggest that G1 N-terminal trimerization
‘occurs co-translationally, with nascent chains interacting
‘with each other when they have traversed past the midpoint
cf the polysome, and that this process is ATP independent.
Results
Oligomeric status of newly synthesized
fulhlength ot
We haye established previously an in vitro ol translation
system to elucidate part of the G1 folding pathway and
2652
A B [A/B]
1 20 35545 5
Fig. 2, Analysis of ia vitro co-ranslation produets of full-length (A)
and truncated (a4) (B) SU wranscripts at various Eoneentations,
The two transcripts were translated separate, or mine (te
Tati) and sarious dilutions ofthe mixture were trslated. Rest
‘were stopped after 60 min and samples were analyzed by SDS-PAGE
‘under not-dssociating conditions (37°C preincubation). Electr
phoresis conditions were such thatthe gl was run for w further 5 h
lafler the dye front had reached the botlom of the gel. Reactions in
lanes and 2 contained 200 ng of RNA each. Reactions in lanes 3-3
‘contained 1.2 ug, 400 ng and 133 ng. wespectively, of I: mature of
FL an 4 RNA, All the lanes were from the same gel, However,
‘due tothe ciferent amounts of RNA wsed for traslation, different
‘exposure times were used sch that each fane shown had
pprovimately the same relative protein band intensities i.e lanes 1
sn 2 were exposed for 36 hand lanes 3-5 were exposed for 12.36
And 108, respectively),
have shown that the N- and C-terminal halves of 61 each
possesses a distinct trimerization domain (Leone et al.,
1992). To determine the timing of the initial trimerization
event (at the N-terminus) relative to translation, we first
‘examined the oligomeric status of newly synthesized full-
length (FL) ol, To this end, FL $1 transcripts encoding
I were translated in rabbit reticulocyte lysates for 7 min
Polysomes were then removed by ultracentrifugation at
4°C and o1 products in the supernatant were chased
further for 60 min at 37°C. Pulse and chase samples were
analyzed by SDS-PAGE under non-dissociating conditions,
that allowed for the detection of stable trimeric 61 forms
(stabilized via the formation of the three-stranded o-helical
coiled-coil at the N-terminus). The results (Figure 1)
show that, immediately after release from polysomes, 61
migrated as monomers (49 kDa molecular weight) under
the conditions used (lane 1). However, a significant
population could eventually be chased into the SDS-stable
Uwimeric form (lane 3). Essentially the same results were
obtained using a trincated 61 with 44 amino acids deleted
from the C-terminus (lanes 2 and 4), We can therefore
conclude that stable N-terminal trimer formation occurs
posttranslationally,
It was necessary to determine whether the newly
released 61 polypeptides were true monomers (in which
case the trimerization mentioned above would be a de facto
post-translational event) or simply unstable (SDS-sens-
itive) 1 trimer complexes that were formed during
translation, and that were converted to the SDS-stable
form upon subsequent chase. ‘To this end, approximately
equimolar amounts of transcripts coding for FL (A) and
the C-terminally truncated product (B) were co-translated
at various, but proportionately equal, RNA concentrations,
and the products analyzed by SDS-PAGE under non-
dissociating conditions. It was rationalized that if the
Reovirus protein «1 trimerization
Nondonaturing SDS-PAGE
PrP eT 2
= monomer
\— Peak Fractions —
@i-27)
Donaturing SOS-PAGE
E
<
“ a <—— Prak Fractions —
Fraction Number @t-27)
Fig. 3. Detection of polysome-bound trimers. (A} Sedimentation profile of polysomes from an in vitwo G1 translation reaction. Protein of was
tuansated in vitro for 10 min and the reaction mixture was subjected (0 eenfugation through a 10-43% sucrose gradient as described in Materials
and methoss. The arrow indicates the direction of sedimentation. Gradient fractions were analyzed for absorbance st 254 oyn. (B) SDS-PAGE af!
peak polysomal fractions. Peak fractions were subjeted to non denaiuring ot denaturing SDS-PAGE
resins in protein sample bufer Jor 30 min at °C, followed by SDS-PAG
Deiled for 5 min in protein sample buffer prior to SDS-PAGE at ron temperate, The kflmos lane in each gel contained
Prior to sucrose eraent centlusation,
newly released chains were truly monomeric, A and B 31
products would assort randomly to generate the four
trimeric species Ay, AxBj, A,B) and B, in the ratio of
1:3:3:1, Furthermore, this ratio would not change with the
proportionate reduction of the concentrations of the wo
transcripts in the reaction mixture, If, on the other hand,
trimeric structures (or structures poised to trimerize) were
formed co-translationally, then the predominant species
formed would be the homotrimers A, and B,, Formation
of the heterotrimers AB) and A,B2 would still be possible
at high transcript concentrations due to the interaction
between the N-termini of nascent GI chains from adjacent
transcripts. At low transcript concentrations, however, the
formation of such heterotrimers would be expected to be
affected drastically relative to that of homotrimers. The
results of such an experiment are presented in Figure 2.
Even at high transcript concentration (40 jig/ml, the
distribution of the four trimeric species (A3, A2B,, AjB;
‘and By) was not in the ratio of 1:3:3:1 as would be
expected had assortment occurred randomly (lane 3),
‘The observed ratio of ~I:I:1:1 suggests a preference of
homoirimer over heterotrimer formation. Importantly, this
1 non-denaturing SDS-PAGE, samples were
lo cared out a 4°C. For denaturing SDS-PAGE. samples were
bias was amplified drastically when the amounts of the
two RNA transcripts were reduced proportionately (lanes
4 and 5), These results strongly argue for the notion
that nascent G1 chains are recruited predominantly co-
‘ranslationally in unstable complexes, and that the chains
within these complexes are committed, upon their release
from polysomes, to form stable trimers (via the
Neterminas).
Detection of trimeric 01 forms on polysomes
In an attempt to demonstrate that o1 nascent chains indeed
form trimers while on polysomes, reactions containing
translated FL transcripts were subjected to suerose gradient
centrifugation (Figure 3A), Fractions corresponding to the
well-resolved polysomal peak were then analyzed directly
by SDS-PAGE under conditions that were less stringent
than those used in Figure 1 (i.e. pre-incubation of samples
in protein sample buffer at 4°C rather than 37°C, followed
by electrophoresis carried out at 4°C). The results (Figure
3B) show that, in addition to 61 monomers, 61 timers
were also present. These trimers would represent the
homotrimers whose assembly was favored in the co-
2653
Vous aimerez peut-être aussi
- The Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeD'EverandThe Subtle Art of Not Giving a F*ck: A Counterintuitive Approach to Living a Good LifeÉvaluation : 4 sur 5 étoiles4/5 (5794)
- Список обзоровDocument5 pagesСписок обзоровapi-3706027Pas encore d'évaluation
- The Little Book of Hygge: Danish Secrets to Happy LivingD'EverandThe Little Book of Hygge: Danish Secrets to Happy LivingÉvaluation : 3.5 sur 5 étoiles3.5/5 (400)
- 18 Hartl TriggerDocument11 pages18 Hartl Triggerapi-3706027Pas encore d'évaluation
- Shoe Dog: A Memoir by the Creator of NikeD'EverandShoe Dog: A Memoir by the Creator of NikeÉvaluation : 4.5 sur 5 étoiles4.5/5 (537)
- Article 15 PresentationDocument10 pagesArticle 15 Presentationapi-3706027Pas encore d'évaluation
- Hidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceD'EverandHidden Figures: The American Dream and the Untold Story of the Black Women Mathematicians Who Helped Win the Space RaceÉvaluation : 4 sur 5 étoiles4/5 (895)
- 23 NacDocument7 pages23 Nacapi-3706027Pas encore d'évaluation
- The Yellow House: A Memoir (2019 National Book Award Winner)D'EverandThe Yellow House: A Memoir (2019 National Book Award Winner)Évaluation : 4 sur 5 étoiles4/5 (98)
- 19 SRPDocument6 pages19 SRPapi-3706027Pas encore d'évaluation
- The Emperor of All Maladies: A Biography of CancerD'EverandThe Emperor of All Maladies: A Biography of CancerÉvaluation : 4.5 sur 5 étoiles4.5/5 (271)
- 20 DaviesDocument4 pages20 Daviesapi-3706027Pas encore d'évaluation
- A Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryD'EverandA Heartbreaking Work Of Staggering Genius: A Memoir Based on a True StoryÉvaluation : 3.5 sur 5 étoiles3.5/5 (231)
- 22 BipDocument12 pages22 Bipapi-3706027Pas encore d'évaluation
- Never Split the Difference: Negotiating As If Your Life Depended On ItD'EverandNever Split the Difference: Negotiating As If Your Life Depended On ItÉvaluation : 4.5 sur 5 étoiles4.5/5 (838)
- 16 NFKBDocument10 pages16 NFKBapi-3706027Pas encore d'évaluation
- Grit: The Power of Passion and PerseveranceD'EverandGrit: The Power of Passion and PerseveranceÉvaluation : 4 sur 5 étoiles4/5 (588)
- 11 GapDocument10 pages11 Gapapi-3706027Pas encore d'évaluation
- On Fire: The (Burning) Case for a Green New DealD'EverandOn Fire: The (Burning) Case for a Green New DealÉvaluation : 4 sur 5 étoiles4/5 (74)
- 12 tRNAmimicryDocument7 pages12 tRNAmimicryapi-3706027100% (1)
- 14 GFPDocument6 pages14 GFPapi-3706027Pas encore d'évaluation
- Elon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureD'EverandElon Musk: Tesla, SpaceX, and the Quest for a Fantastic FutureÉvaluation : 4.5 sur 5 étoiles4.5/5 (474)
- 17 NicolaDocument5 pages17 Nicolaapi-3706027Pas encore d'évaluation
- Devil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaD'EverandDevil in the Grove: Thurgood Marshall, the Groveland Boys, and the Dawn of a New AmericaÉvaluation : 4.5 sur 5 étoiles4.5/5 (266)
- 09 TM RnaDocument6 pages09 TM Rnaapi-3706027Pas encore d'évaluation
- The Unwinding: An Inner History of the New AmericaD'EverandThe Unwinding: An Inner History of the New AmericaÉvaluation : 4 sur 5 étoiles4/5 (45)
- 07 ChapevilleDocument7 pages07 Chapevilleapi-3706027Pas encore d'évaluation
- 02 Ogata NoharaDocument2 pages02 Ogata Noharaapi-3706027Pas encore d'évaluation
- Team of Rivals: The Political Genius of Abraham LincolnD'EverandTeam of Rivals: The Political Genius of Abraham LincolnÉvaluation : 4.5 sur 5 étoiles4.5/5 (234)
- Synth Pro11Document1 pageSynth Pro11api-3706027Pas encore d'évaluation
- 04 CrickDocument6 pages04 Crickapi-3706027Pas encore d'évaluation
- 06 Brenner JacobDocument6 pages06 Brenner Jacobapi-3706027Pas encore d'évaluation
- The World Is Flat 3.0: A Brief History of the Twenty-first CenturyD'EverandThe World Is Flat 3.0: A Brief History of the Twenty-first CenturyÉvaluation : 3.5 sur 5 étoiles3.5/5 (2259)
- 05 Gros WatsonDocument5 pages05 Gros Watsonapi-3706027Pas encore d'évaluation
- The Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreD'EverandThe Gifts of Imperfection: Let Go of Who You Think You're Supposed to Be and Embrace Who You AreÉvaluation : 4 sur 5 étoiles4/5 (1090)
- 01 SpirinDocument2 pages01 Spirinapi-3706027Pas encore d'évaluation
- Synth Pro11Document2 pagesSynth Pro11api-3706027Pas encore d'évaluation
- The Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersD'EverandThe Hard Thing About Hard Things: Building a Business When There Are No Easy AnswersÉvaluation : 4.5 sur 5 étoiles4.5/5 (344)
- Synth Pro11Document1 pageSynth Pro11api-3706027Pas encore d'évaluation
- Synth Pro11Document1 pageSynth Pro11api-3706027Pas encore d'évaluation
- Synth Pro10Document1 pageSynth Pro10api-3706027Pas encore d'évaluation
- Synth Pro10Document2 pagesSynth Pro10api-3706027Pas encore d'évaluation
- Synth Pro10Document1 pageSynth Pro10api-3706027Pas encore d'évaluation
- Synth Pro10Document1 pageSynth Pro10api-3706027Pas encore d'évaluation
- Synth Pro3Document3 pagesSynth Pro3api-3706027Pas encore d'évaluation
- The Sympathizer: A Novel (Pulitzer Prize for Fiction)D'EverandThe Sympathizer: A Novel (Pulitzer Prize for Fiction)Évaluation : 4.5 sur 5 étoiles4.5/5 (121)
- Synth Pro3Document2 pagesSynth Pro3api-3706027Pas encore d'évaluation
- 003 REVIEWER IN SCIENCE 7-SUgarDocument3 pages003 REVIEWER IN SCIENCE 7-SUgarrizza suarezPas encore d'évaluation
- WWW - Manabadi.co - in Timetables Svubsc07032009Document6 pagesWWW - Manabadi.co - in Timetables Svubsc07032009Lokesh ModemzPas encore d'évaluation
- 8th Class Biology Project WorkDocument10 pages8th Class Biology Project WorkPRANU83% (6)
- Water PotentialDocument35 pagesWater PotentialLuis GarcíaPas encore d'évaluation
- PNU Final Coaching Gen - EdDocument15 pagesPNU Final Coaching Gen - EdTatinPas encore d'évaluation
- CS 2000iDocument88 pagesCS 2000iBintang MaulanaPas encore d'évaluation
- SBRC HematologyOncology 2Document80 pagesSBRC HematologyOncology 2dalia khamoPas encore d'évaluation
- Health Status and Quality of Life of Geriatric Population in Old Age Homes and Living With Family in Chennai A Comparative StudyDocument5 pagesHealth Status and Quality of Life of Geriatric Population in Old Age Homes and Living With Family in Chennai A Comparative StudyInternational Journal of Innovative Science and Research TechnologyPas encore d'évaluation
- Ujmr 2020 - 4 - 24 - 30 PDFDocument7 pagesUjmr 2020 - 4 - 24 - 30 PDFUMYU Journal of Microbiology Research (UJMR)Pas encore d'évaluation
- Silviculture: February 2018Document69 pagesSilviculture: February 2018Tang YinPas encore d'évaluation
- Her Body and Other Parties: StoriesD'EverandHer Body and Other Parties: StoriesÉvaluation : 4 sur 5 étoiles4/5 (821)
- Induction Presentation Biology A Level and Gcse.204719226Document25 pagesInduction Presentation Biology A Level and Gcse.204719226Cut TirayaPas encore d'évaluation
- Applied Zoology: Pearl Culture Bechan LalDocument16 pagesApplied Zoology: Pearl Culture Bechan LalSk RajPas encore d'évaluation
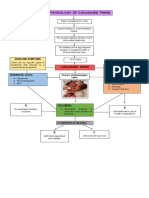
- Pathophysiology of Conjoined TwinsDocument1 pagePathophysiology of Conjoined TwinsPrincess Loreto AlegrePas encore d'évaluation
- Answers For Support Worksheet - Option F: 1 A Feature Bacteria Archaea EukaryotaDocument1 pageAnswers For Support Worksheet - Option F: 1 A Feature Bacteria Archaea EukaryotaHellö PëachPas encore d'évaluation
- 2457-Article Text-14907-2-10-20120724Document6 pages2457-Article Text-14907-2-10-20120724desi meleniaPas encore d'évaluation
- Cse Integration Lesson PlanDocument5 pagesCse Integration Lesson PlanCarl Anthony Lague PahuyoPas encore d'évaluation
- Strategies For Plant Regeneration From Mesophyll Protoplasts of The Recalcitrant Fruit and Farmwoodland Species L. (Sweet/wild Cherry)Document6 pagesStrategies For Plant Regeneration From Mesophyll Protoplasts of The Recalcitrant Fruit and Farmwoodland Species L. (Sweet/wild Cherry)Geral BlancoPas encore d'évaluation
- MFK-Drug Management CycleDocument32 pagesMFK-Drug Management CycleLhynda Priarti Latif100% (1)
- Anima and Animus Term ReportDocument17 pagesAnima and Animus Term ReportSadia Shahid100% (1)
- Unit 3Document61 pagesUnit 3JessicaHaePas encore d'évaluation
- Characteristics of A Community PDFDocument15 pagesCharacteristics of A Community PDFAlijanPas encore d'évaluation
- Lesson 3.1 History of Life On EarthDocument1 pageLesson 3.1 History of Life On EarthJoshua EboraPas encore d'évaluation
- Lactic Acid Production From Cane MolassesDocument4 pagesLactic Acid Production From Cane MolassesSenkatuuka LukePas encore d'évaluation
- Hubungan Tingkat Pengetahuan Dan Dukungan Keluarga Terhadap Kepatuhan Ibu Hamil Mengkonsumsi Tablet Zat BesiDocument13 pagesHubungan Tingkat Pengetahuan Dan Dukungan Keluarga Terhadap Kepatuhan Ibu Hamil Mengkonsumsi Tablet Zat Besinovilia eka sariPas encore d'évaluation
- Upcoming Predicted Material For PTE-A Exam Feb-2019Document123 pagesUpcoming Predicted Material For PTE-A Exam Feb-2019GauravPas encore d'évaluation
- Aspb 2012 - 106Document204 pagesAspb 2012 - 106arunprabhu_dhanapalPas encore d'évaluation
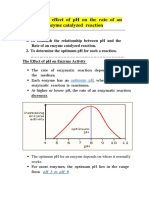
- Effect of PH On Enzyme ActivityDocument12 pagesEffect of PH On Enzyme ActivityAb AbPas encore d'évaluation
- Treatment of Latent Tuberculosis InfectionDocument10 pagesTreatment of Latent Tuberculosis InfectionRichard WoolitePas encore d'évaluation
- A Brief Introduction To ArgumentsDocument7 pagesA Brief Introduction To ArgumentsJohn DarwinPas encore d'évaluation
- (English (Auto-Generated) ) Ep. 94 - Andres Ryan of BioAquatix - Com On Breeding Clown Loaches (DownSub - Com)Document78 pages(English (Auto-Generated) ) Ep. 94 - Andres Ryan of BioAquatix - Com On Breeding Clown Loaches (DownSub - Com)acromontiPas encore d'évaluation