Académique Documents
Professionnel Documents
Culture Documents
Drosophila: The Reaction and Inhibition Mechanisms of Alcohol Dehydrogenase
Transféré par
runjuythDescription originale:
Titre original
Copyright
Formats disponibles
Partager ce document
Partager ou intégrer le document
Avez-vous trouvé ce document utile ?
Ce contenu est-il inapproprié ?
Signaler ce documentDroits d'auteur :
Formats disponibles
Drosophila: The Reaction and Inhibition Mechanisms of Alcohol Dehydrogenase
Transféré par
runjuythDroits d'auteur :
Formats disponibles
The reaction and inhibition mechanisms of
Drosophila alcohol dehydrogenase
J. Benach, R. Meijers , S. Atrian , R. Gonzlez-Duarte , V. Lamzin & R. Ladenstein Center for Structural Biochemistry, Karolinska Institute, S-141 57 Huddinge, Sweden EMBL c/o DESY, Notkestrasse 85 Geb. 25a, D-22603 Hamburg, Germany Department of Genetics, University of Barcelona, Av. Diagonal 645, 08028 Barcelona, Spain Project number: PX-99-362 Beamline: X11
1 2 2 1
1 2
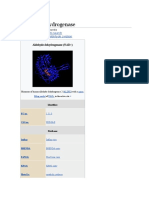
Drosophila alcohol dehydrogenase (DADH; EC 1.1.1.1) is an NAD(H)-dependent oxidoreductase that catalyzes the oxidation of alcohols to aldehydes/ketones. This enzyme is one of the most well characterized member of the superfamily of the short-chain dehydrogenases/reductases (SDR) [1]. DADH subunits show an / single domain structure with a characteristic NAD(H) binding motif (Rossmann fold) [2]. The active site pocket comprises a hydrophobic and bifurcated cavity that explains why the enzyme is more efficient in oxidizing secondary aliphatic alcohols -preferably R enantiomers- than primary ones. High resolution studies on the 3-pentanone/NAD+/enzyme complex showed that the ketone inhibitor molecule has undergone a covalent reaction with the coenzyme in the ternary complex [3,4].
Figure 1: (a) Stereoview of the 3-pentanone-NAD+ adduct observed at 1.4 resolution and (b) zoomed section showing the covalent interaction between the ketone and the coenzyme.
Due to the presence of the positively charged ring of the coenzyme (NAD+) and the residue Lys155, the amino acid Tyr151 is in its deprotonated state at physiological pH. Tyr151 can abstract a proton from the enolic form of the ketone and catalyze a nucleophilic attack of the C atom to the C4 position of the coenzyme creating an NAD-ketone adduct. The binding of these NAD-ketone adducts to DADH accounts for the inactivation of the enzyme. In order to further understand the catalytic reaction mechanism of DADH crystals of a ternary complex of this enzyme and NAD+ and trifluoroethanol were obtained. Data was measured at the X11 beamline at the Hamburg outstation and diffracted beyond 1.1 resolution. The crystal structure of the ternary complex is still undergoing crystallographic refinement.
Figure 2: Zoomed section showing the active site of the ternary complex of Drosophila alcohol dehydrogenase at 1.1 resolution showing the main amino acids involved in catalytic activity (Ser138, Tyr151, Lys155), the NAD+ and the trifluoroethanol molecules. 3Fo-2Fc electron density map contoured at 1.7 and 4.4.
Table 1. X-ray data collection statistics for DADH trifluoroethanol NAD+ ternary complex
Cell parameters Space group Total reflections Unique reflections Resolution range Completeness R-merge I/sigma > 2 Multiplicity >2 Source Wavelength Temperature a=66.30 , b=52.73 , c=69.92 , =107.38 deg. P21 3758910 195445 50-1.05 (1.14 -1.12 ) 92.0% (85.2 %) 3.7% (32.7%) 74.6% (55.5%) 76% (72%) X11 EMBL, Hamburg 0.9092 100 K
References
[1] H. Jrnvall, B. Persson, M. Krook, S. Atrian, R. Gonzlez-Duarte, J. Jeffery, and D. Ghosh, Biochemistry, 34, 6003 (1995) [2] J. Benach, S. Atrian, R. Gonzlez-Duarte, and R. Ladenstein, J. Mol. Biol. 282, 383 (1998) [3] J. Benach, S. Atrian, R. Gonzlez-Duarte, and R. Ladenstein, J. Mol. Biol. 289, 335 (1999) [4] J. Benach. Doctoral thesis. Karolinska Institute. Stockholm, Sweden. (1999)
Vous aimerez peut-être aussi
- 13 Musculoskeletal System - ATFDocument20 pages13 Musculoskeletal System - ATFipek terimPas encore d'évaluation
- DrDerm Devices EnglishDocument60 pagesDrDerm Devices EnglishAdrian RosuPas encore d'évaluation
- Biomechanics in Applications PDFDocument424 pagesBiomechanics in Applications PDFRAUL EDUARDO GUTIERREZ COITIÑO100% (1)
- Digestion Test BiologyDocument3 pagesDigestion Test BiologyKatrīna SimanovskaPas encore d'évaluation
- Tingling and NumbnessDocument67 pagesTingling and NumbnessAradhanaRamchandaniPas encore d'évaluation
- Assessment Nursing Diagnosis Analysis Goals and Objectives Nursing Interventions Rationale Evaluation EffectivenessDocument3 pagesAssessment Nursing Diagnosis Analysis Goals and Objectives Nursing Interventions Rationale Evaluation EffectivenessYnah Sayoc75% (4)
- Good BooksDocument16 pagesGood BooksFbihansip bancel100% (1)
- NCPsDocument13 pagesNCPsRocel DevillesPas encore d'évaluation
- Antigen & AntibodiesDocument16 pagesAntigen & AntibodiesYing Ming TangPas encore d'évaluation
- Neural EngineeringDocument377 pagesNeural Engineeringbeesah100% (1)
- Global Atlas of AsthmaDocument196 pagesGlobal Atlas of AsthmaMinerva Stanciu50% (2)
- Anatomy & Physiology of Olfactory System.Document27 pagesAnatomy & Physiology of Olfactory System.Prasanna DattaPas encore d'évaluation
- Functional Electrospun Fibrous Scaffolds With Dextran-G-Poly (L-Lysine) - Vapg/Microrna-145 To Specially Modulate Vascular SmcsDocument9 pagesFunctional Electrospun Fibrous Scaffolds With Dextran-G-Poly (L-Lysine) - Vapg/Microrna-145 To Specially Modulate Vascular Smcssyedamasoomazahra9Pas encore d'évaluation
- Aldehyde Dehydrogenase: Jump To Navigation Jump To SearchDocument11 pagesAldehyde Dehydrogenase: Jump To Navigation Jump To SearchChaeyoung SonPas encore d'évaluation
- Takahiro Inui, Hisafumi Ikeda and Yushin Nakamura - Design of An Artificial Restriction Enzyme Having Simultaneous DNA Cleavage ActivityDocument2 pagesTakahiro Inui, Hisafumi Ikeda and Yushin Nakamura - Design of An Artificial Restriction Enzyme Having Simultaneous DNA Cleavage ActivityGmso3Pas encore d'évaluation
- Benzodiazepain Like ActionDocument6 pagesBenzodiazepain Like ActionMohamed KhedrPas encore d'évaluation
- Denaturasi DnaDocument8 pagesDenaturasi DnadetiPas encore d'évaluation
- Ye 2000Document8 pagesYe 2000h.sinner671Pas encore d'évaluation
- Divate Et Al. - 2017 - Metabolic Engineering of Isaccharomyces Cerevisiaei For Improvement in Stresses Tolerance-AnnotatedDocument13 pagesDivate Et Al. - 2017 - Metabolic Engineering of Isaccharomyces Cerevisiaei For Improvement in Stresses Tolerance-AnnotatedIdier MoRoPas encore d'évaluation
- Understanding Retinol Metabolism: Structure and Function of Retinol DehydrogenasesDocument4 pagesUnderstanding Retinol Metabolism: Structure and Function of Retinol DehydrogenasesSergio OrtoPas encore d'évaluation
- PantDocument16 pagesPantJili BiliPas encore d'évaluation
- European Journal of Biochemistry - 2002 - Cherepanov - Dynamic Mechanism of Nick Recognition by DNA LigaseDocument7 pagesEuropean Journal of Biochemistry - 2002 - Cherepanov - Dynamic Mechanism of Nick Recognition by DNA LigaseAnuvansh SinghPas encore d'évaluation
- Nucl. Acids Res. 2002 KitDocument6 pagesNucl. Acids Res. 2002 KitAbdul Mueez LonePas encore d'évaluation
- Article MoleculesDocument15 pagesArticle MoleculesDenisaPas encore d'évaluation
- Enfoque Ambiental Xu2014 Article DioxinAndDioxin-LikeCompoundsSDocument7 pagesEnfoque Ambiental Xu2014 Article DioxinAndDioxin-LikeCompoundsSJefersson TiquePas encore d'évaluation
- Kotov 2014 EnglDocument13 pagesKotov 2014 EnglLudmila OleninaPas encore d'évaluation
- Rosales 2008Document4 pagesRosales 2008Ezequiel SaavedraPas encore d'évaluation
- 1,5-Dichloroethanoanthracene Derivatives As AntidepressantDocument11 pages1,5-Dichloroethanoanthracene Derivatives As Antidepressantمجيب سلطانPas encore d'évaluation
- 1989 Hanukoglu Gutfinger European Journal of Biochemistry FEBS 180 2 cDNA Sequence of Adrenodoxin ReductaseDocument6 pages1989 Hanukoglu Gutfinger European Journal of Biochemistry FEBS 180 2 cDNA Sequence of Adrenodoxin Reductasel4vfeaokf5Pas encore d'évaluation
- High-Performance Liquid Chromatographic Separation and Chiroptical Properties of The Enantiomers of Naringenin and Other FlavanonesDocument8 pagesHigh-Performance Liquid Chromatographic Separation and Chiroptical Properties of The Enantiomers of Naringenin and Other Flavanonesfernando gonzalezPas encore d'évaluation
- Extracellular Superoxide Dismutase (SOD3) : An Antioxidant or Prooxidant in The Extracellular Space?Document33 pagesExtracellular Superoxide Dismutase (SOD3) : An Antioxidant or Prooxidant in The Extracellular Space?Khairul AsriPas encore d'évaluation
- Nicotinamide Adenine Dinucleotide - WikipediaDocument116 pagesNicotinamide Adenine Dinucleotide - WikipediaBashiir NuurPas encore d'évaluation
- Science Glycerol 5Document11 pagesScience Glycerol 5Iju IzzuPas encore d'évaluation
- Cloning, Purification and Comparative Characterization of Two Digestive Lysozymes From Musca Domestica LarvaeDocument9 pagesCloning, Purification and Comparative Characterization of Two Digestive Lysozymes From Musca Domestica LarvaebobyjuniorPas encore d'évaluation
- Functional Domains of An ATP-dependent DNA Ligase: Aidan J. Doherty and Dale B. WigleyDocument9 pagesFunctional Domains of An ATP-dependent DNA Ligase: Aidan J. Doherty and Dale B. WigleyLathoifulIsyarohPas encore d'évaluation
- 檔案下載Document6 pages檔案下載Rosalina DjatmikaPas encore d'évaluation
- 17Document9 pages17'Licenza AdagioPas encore d'évaluation
- Size Effect of Layered Double Hydroxide Platelets On The Crystallization Behavior of Isotactic PolypropyleneDocument8 pagesSize Effect of Layered Double Hydroxide Platelets On The Crystallization Behavior of Isotactic PolypropyleneaegosmithPas encore d'évaluation
- Proteolytic Dissection of Zab, The Z-DNA-binding Domain of Human ADAR1Document8 pagesProteolytic Dissection of Zab, The Z-DNA-binding Domain of Human ADAR1Saradindu GhoshPas encore d'évaluation
- Eur. J, 2010, 16, 6509-6517 Reek Anti-HalpernDocument9 pagesEur. J, 2010, 16, 6509-6517 Reek Anti-HalpernszbaloghPas encore d'évaluation
- Jurnal PenuntunDocument11 pagesJurnal PenuntunAgustinus PotterPas encore d'évaluation
- Candida Tropicalis: Characterization of Xylose Reductase From Immobilized On Chitosan BeadDocument12 pagesCandida Tropicalis: Characterization of Xylose Reductase From Immobilized On Chitosan BeadkashvinwarmaPas encore d'évaluation
- Le Chauhan 2014 Simple and Short Synthesis of Trans R Nerolidol A Pheromone Component of Fruit Spotting BugDocument2 pagesLe Chauhan 2014 Simple and Short Synthesis of Trans R Nerolidol A Pheromone Component of Fruit Spotting BugotpmairieuhihiPas encore d'évaluation
- Inmovilización de LDHDocument12 pagesInmovilización de LDHjessicrmPas encore d'évaluation
- ACIE, Volume 48, Issue 41, Pages 7577-7581, 2009Document5 pagesACIE, Volume 48, Issue 41, Pages 7577-7581, 2009John TanPas encore d'évaluation
- Vitae 0121-4004: Issn: Vitae@udea - Edu.coDocument9 pagesVitae 0121-4004: Issn: Vitae@udea - Edu.coDaniela GonzálezPas encore d'évaluation
- Benzopyrazines: Synthesis, Characterization and Evaluation As Aldose Reductase InhibitorsDocument8 pagesBenzopyrazines: Synthesis, Characterization and Evaluation As Aldose Reductase InhibitorsWalid Ebid ElgammalPas encore d'évaluation
- Hydrogenation of Naphthalene and Methylnaphthalene: Modeling and SpectrosDocument12 pagesHydrogenation of Naphthalene and Methylnaphthalene: Modeling and Spectrosioanaandra5690Pas encore d'évaluation
- Santoro Et Al-2008-Advanced Synthesis & CatalysisDocument4 pagesSantoro Et Al-2008-Advanced Synthesis & CatalysisIsa Guerrero TroyanoPas encore d'évaluation
- Van Acker, 1996Document8 pagesVan Acker, 1996Sergio mauricio sergioPas encore d'évaluation
- Applied-Determination of The Binding Constant-Latona D FDocument4 pagesApplied-Determination of The Binding Constant-Latona D FImpact JournalsPas encore d'évaluation
- A Study of The Kinetics and Mechanism of Yeast Alcohol Dehydrogenase - Dickinson & Monger 1972Document10 pagesA Study of The Kinetics and Mechanism of Yeast Alcohol Dehydrogenase - Dickinson & Monger 1972MeidayPas encore d'évaluation
- Articulo 10.-Boric Acid As A Green Catalyst For The Conversion of Aldehydes and KetonesDocument8 pagesArticulo 10.-Boric Acid As A Green Catalyst For The Conversion of Aldehydes and KetonesPaul Delgado MendozaPas encore d'évaluation
- Asian J. Org. Chem. 2015, 4, 28 - 32 PDFDocument5 pagesAsian J. Org. Chem. 2015, 4, 28 - 32 PDFSulagna DasPas encore d'évaluation
- Fluid Phase Equilibria: Wen-Cai Wang, Yang-Yang Zhang-Bian, Guo-Liang Zhang, Qing Xia, Feng-Bao ZhangDocument5 pagesFluid Phase Equilibria: Wen-Cai Wang, Yang-Yang Zhang-Bian, Guo-Liang Zhang, Qing Xia, Feng-Bao ZhangMuhammad Abdur RokhimPas encore d'évaluation
- European Journal of Inorganic Chemistry, 2010, 4187-4195.Document9 pagesEuropean Journal of Inorganic Chemistry, 2010, 4187-4195.Pradhumn SinghPas encore d'évaluation
- LDH Based NanocompositesDocument10 pagesLDH Based NanocompositesDrHassam MazharPas encore d'évaluation
- Chen 2011Document5 pagesChen 2011Mario RojasPas encore d'évaluation
- Olga Ferreira (2012) - Solubility of Flavonoids in Pure SolventsDocument5 pagesOlga Ferreira (2012) - Solubility of Flavonoids in Pure SolventsDAVID DUARTEPas encore d'évaluation
- A General Method For C Reductive Alkylation of Indoles: Anu Mahadevan, Howard Sard, Mario Gonzalez and John C. MckewDocument3 pagesA General Method For C Reductive Alkylation of Indoles: Anu Mahadevan, Howard Sard, Mario Gonzalez and John C. MckewMichell UrzúaPas encore d'évaluation
- 8-PNBD - Applied Polymer ScienceDocument6 pages8-PNBD - Applied Polymer ScienceDiêgo MedeirosPas encore d'évaluation
- Clivagem Cobre SufatoDocument5 pagesClivagem Cobre SufatoVARAL CIÊNCIAPas encore d'évaluation
- Matthew AdamsDocument3 pagesMatthew AdamsKathy T. Adams100% (1)
- Glyconanoparticles Activity 3Document3 pagesGlyconanoparticles Activity 3ЛуизАпазаТ.Pas encore d'évaluation
- J. Biol. Chem.-2003-Ayyagari-1618-25Document8 pagesJ. Biol. Chem.-2003-Ayyagari-1618-25A. M. Mahedi HasanPas encore d'évaluation
- Water Vs Solvent Free ConditionDocument18 pagesWater Vs Solvent Free Conditionintata 24Pas encore d'évaluation
- RodioDocument5 pagesRodioTavo RodriguezPas encore d'évaluation
- Keap1 Recruits Neh2 Through Binding To ETGE and DLG Motifs: Characterization of The Two-Site Molecular Recognition ModelDocument14 pagesKeap1 Recruits Neh2 Through Binding To ETGE and DLG Motifs: Characterization of The Two-Site Molecular Recognition Model1766636545Pas encore d'évaluation
- Ligases: Rachel Nash and Tomas LindahlDocument12 pagesLigases: Rachel Nash and Tomas Lindahlss9194Pas encore d'évaluation
- Angela Seidl and Hans-Jurgen Hinz - The Free Energy of DNA Supercoiling Is Enthalpy-DeterminedDocument5 pagesAngela Seidl and Hans-Jurgen Hinz - The Free Energy of DNA Supercoiling Is Enthalpy-DeterminedDopamePas encore d'évaluation
- 1 s2.0 S0008621503004713 MainDocument9 pages1 s2.0 S0008621503004713 MainJoão VictorPas encore d'évaluation
- In Brief: Inguinal HerniaDocument4 pagesIn Brief: Inguinal HerniaSaf DicamPas encore d'évaluation
- ANS & CVS Response To Exercise v2Document18 pagesANS & CVS Response To Exercise v2chow wing yin amandaPas encore d'évaluation
- Perfusionist Job DescriptionDocument3 pagesPerfusionist Job DescriptionNouman IshaqPas encore d'évaluation
- Bds SyllabusDocument67 pagesBds Syllabusabdulbasitejaz100% (2)
- Rethinking Tongue Tie Anatomy - Anterior Vs Posterior Is Irrelevant - DrGhaheriDocument4 pagesRethinking Tongue Tie Anatomy - Anterior Vs Posterior Is Irrelevant - DrGhaheriKumaran Bagavathi RagavanPas encore d'évaluation
- A Rare Variation of The Digastric Muscle: Original ResearchDocument3 pagesA Rare Variation of The Digastric Muscle: Original ResearchYeraldin EspañaPas encore d'évaluation
- List of Medical Roots, Suffixes and Prefixes - Wikipedia, The Free EncyclopediaDocument35 pagesList of Medical Roots, Suffixes and Prefixes - Wikipedia, The Free EncyclopediaHoward GouldPas encore d'évaluation
- Cell and Its Structure and Functions: Q1. Write Down The Main Differences Between The Animal Cell and The Plant CellDocument5 pagesCell and Its Structure and Functions: Q1. Write Down The Main Differences Between The Animal Cell and The Plant CellMuhhammed AliPas encore d'évaluation
- 0046 8177 (93) 90013 7Document12 pages0046 8177 (93) 90013 7Carmen Gomez MuñozPas encore d'évaluation
- Sbk3023 Food Science and NutritionDocument20 pagesSbk3023 Food Science and NutritionKuMohdSyafiqPas encore d'évaluation
- EOT2 BiologyDocument21 pagesEOT2 BiologyKasumi SatoPas encore d'évaluation
- DT Jan NPR Germain-2 9 FNLDocument8 pagesDT Jan NPR Germain-2 9 FNLKranti PrajapatiPas encore d'évaluation
- Zeev Davidovitch - Electri...Document12 pagesZeev Davidovitch - Electri...klausPas encore d'évaluation
- Anatomy and Physiology of Cornea: DR - Lhacha Wangdi 1st Year Resident Department of Ophthalmology Jdwnrh/KgumsbDocument47 pagesAnatomy and Physiology of Cornea: DR - Lhacha Wangdi 1st Year Resident Department of Ophthalmology Jdwnrh/KgumsbYaman MuhaisenPas encore d'évaluation
- Receptors Ion ChannelsDocument42 pagesReceptors Ion ChannelsMd. Ahsan-Ul BariPas encore d'évaluation
- English IIDocument107 pagesEnglish IIlauroPas encore d'évaluation
- Patel 2014Document7 pagesPatel 2014Javiera Munizaga MuñozPas encore d'évaluation
- 10 Interesting Facts About CatsDocument8 pages10 Interesting Facts About CatsAndreea DanaPas encore d'évaluation