Académique Documents
Professionnel Documents
Culture Documents
Alphaview User Guide
Transféré par
Michael CaverlyCopyright
Formats disponibles
Partager ce document
Partager ou intégrer le document
Avez-vous trouvé ce document utile ?
Ce contenu est-il inapproprié ?
Signaler ce documentDroits d'auteur :
Formats disponibles
Alphaview User Guide
Transféré par
Michael CaverlyDroits d'auteur :
Formats disponibles
AlphaView user guide
This is a brief tutorial on how to use the AlphaView software that is available on the Virtual Campus Website. This tutorial will help you go through the analysis steps you need to do to determine the optimal amount of enzyme to use in your assays next week. There are many other free programs that can do similar analysis (such as ImageJ), so you are not locked into using this program. 1) Install and open the AlphaView software. 2) Open the figure you want to analyse. a. Select the File menu. b. Then, click on Open. c. Choose a picture with a valid format (.tif).
3) Your picture should be fairly straight. You can always rotate your picture by using the rotation tool in the left panel of the screen (Rotate-Flip). You can also slide the Black and White tabs on the left of the screen to adjust the intensity of your figure (do not oversaturate your bands).
1 James Butcher (2011)
4) In some situations, you will need to crop out one series of reactions to analyze. To do this, select Edit Activation under the Edit menu. You can then highlight the section to analyze and click Crop. Make sure you get everything you need in the cropped image. Important Note: There is no undo function in this program. Save frequently and think twice before finalizing a command.
5) To analyse the intensity of our DNA bands, you need to select the Analysis tools tab. Select the Lane profile and then Lane tab. If you scroll down you can see there is a Number of lanes function. Make sure this is set to the right number that you need to analysis. The computer program will now draw some lanes on the gel. Unfortunately, they do not line up with our results; however you can stretch the lanes to fit your gel. (see figures on the following page) You cannot edit the lanes separately with this function (you want to keep everything the same for each lane so this is not a problem).
2 James Butcher (2011)
3 James Butcher (2011)
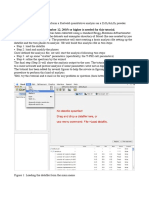
6) Once the lanes fit your sample. You can now click on Detect bands. The program assumes that the bands are lighter than the background. You can change this by selecting the Bands darker than background option if your picture was inverted (white background and black DNA bands). 7) Here the program has detected bands for our gel. The bands are numbered based on their order from the top of the gel. The program also displays a histogram with the peak intensity for each band detected in the selected lane (Lane 1 in this example). Note that two bands visible in Lane 1 were not detected by the software and does not have an intensity associated with them (Red arrows).
8) We can fix this by adding bands. In the Band tab there is an option for adding/deleting bands. Select the Add band function and click on the band you want to analyse. 9) Once all the ladder bands are selected. You can do the same for the remaining lanes. If a visible bands is not selected in a specific lane. Select the lane by clicking on it. Then, select the Add band option and click on the band you want to select (see Lane #2 and 3 on the following pages picture).
4 James Butcher (2011)
The real strength of programs like Alphaview is that they can generate standard curves and calculate densitometry for us. The next few steps will go through this process. 10) Click on the MW tab in Lane profiles. Click on the Add button and click on any ladder band (Its best to start at the top). Now enter in its MW in bp; repeat this for each band visible.
5 James Butcher (2011)
11) The program assumes that the MW migrations are logarithmically related and will generate a standard curve with R2 values as you add MW markers. If your curve isnt great, you can remove outlier bands using the Remove Band button under the Band tab). As you can see on my example, the uppermost band of my marker seems to be an outlier. Let see what will happen if I delete that band (red arrow).
12) This modification improves the curve (R2 of 0.9165 instead of 0.8452), but note that all the other bands have been renumbered because they start from the top. It is important to keep this in mind. You need to keep a copy of this ordering so you remember which band #s in your final report relate to which band on the gel. 13) We can do a similar standard curve generation using masses. Click on the Mass tab and make sure that the option is set to Mass standards. This assumes each band has a known mass. The other option Molecular Ladder assumes that you know the total mass of your ladder, but not of each individual band. Click on Add and insert a mass for each individual band of the marker (see following picture).
6 James Butcher (2011)
14) Now we can export our data. Select the Report tab and make sure the Data Sheet and Graph functions are selected. You should also click on the Full image option (rather than or sized) or the image may be too small to see. Click on the Next button to generate the report. Then press on the Generate report option.
7 James Butcher (2011)
15) First section of your report contains an image of your gel with the bands labelled with their band numbers and any standard curves you generated.
16) The second section is where you will find all the calculated MWs and masses for your bands. Because of the numbering system, the same band in different lanes may have different numbers. Again, this is why you need a copy of the gel image. The green section corresponds to your DNA marker. 17) You can save the report as an HTML file. You can also copy your mass values into excel to test the linearity of the data (see picture on following page). It will be particularly useful when you will need to figure out the best enzyme amount to use in Lab#8.
8 James Butcher (2011)
18) You are now ready to analyse your data. Using excel, you can analyze this data in three different ways, with increasing degrees of complexity, and plotted the data. a. The first is the raw intensity (mass) of the major band present in reach reaction plotted vs time.
180 160 140 Intensity 120 100 80 60 40 20 0 0 5
Intensity vs time
R = 0.9751
10
15 Assay Time
20
25
30
b. The second is the total intensity of enzymatic products for each time point added together and then plotted vs time. If you look carefully at my example, you will notice that in lane 7, a second enzymatic product with a higher molecular weight than the original band becomes visible. If it happens to you, you can add the intensity of both product bands. It will correspond to the total intensity of enzymatic products. 9 James Butcher (2011)
c. The third normalizes the total intensity of enzymatic products for each time point added together, to your loading efficiency in each well. Im not going to tell you how to do this normalization. There is a way to tell your loading efficiency across each lane in your enzymatic reactions due to a factor that should not change across conditions.
5.00 4.50 4.00 3.50 3.00 2.50 2.00 1.50 1.00 0.50 0.00 0
Normalized Total Intensity vs time
Intensity
R = 0.9778
10
15 Assay Time
20
25
30
10 James Butcher (2011)
Vous aimerez peut-être aussi
- Labklass Digital Image AnalysisDocument6 pagesLabklass Digital Image AnalysiscabrahaoPas encore d'évaluation
- Compos 115Document5 pagesCompos 115alexmechdesignerPas encore d'évaluation
- Two Ways To Count Cells With Imagej: Manual Cell Counting and Marking (Plugin Required)Document5 pagesTwo Ways To Count Cells With Imagej: Manual Cell Counting and Marking (Plugin Required)Brunno CaetanoPas encore d'évaluation
- ImageJ Gel AnalysisDocument19 pagesImageJ Gel AnalysisSoledad Zúñiga HernándezPas encore d'évaluation
- Unsupervised Classfication Using ER MapperDocument9 pagesUnsupervised Classfication Using ER MapperavisenicPas encore d'évaluation
- Tutorial For Risa Educational: C.M. Uang and K.M. LeetDocument18 pagesTutorial For Risa Educational: C.M. Uang and K.M. LeetfabianoramiroPas encore d'évaluation
- Track Video Analysis Software TutorialDocument12 pagesTrack Video Analysis Software TutorialdekmepPas encore d'évaluation
- Synthetic Microstructure Tool Generates Representative MicrostructuresDocument5 pagesSynthetic Microstructure Tool Generates Representative MicrostructureschotuPas encore d'évaluation
- Cell Morphology Analysis PDFDocument5 pagesCell Morphology Analysis PDFIon C. AndronachePas encore d'évaluation
- RISA Educational Tutorial for Truss and Frame AnalysisDocument18 pagesRISA Educational Tutorial for Truss and Frame AnalysisAlfredo FranquezPas encore d'évaluation
- Ansys Lab1 PDFDocument15 pagesAnsys Lab1 PDFVenkata DineshPas encore d'évaluation
- PowerMap User Manual v3Document12 pagesPowerMap User Manual v3Alberto UriosteguiPas encore d'évaluation
- Lab 03 - Data Formats - Contrast Stretching - Density SlicingDocument11 pagesLab 03 - Data Formats - Contrast Stretching - Density SlicingANN SHALITAPas encore d'évaluation
- SAP2000 Tutorial by GoelDocument21 pagesSAP2000 Tutorial by GoelhedyPas encore d'évaluation
- Water Table Analysis and VisualizationDocument19 pagesWater Table Analysis and VisualizationDavid Felipe Limas MendigañoPas encore d'évaluation
- Instruction of Using VSoft Viscometer Data Collection, Analysis and Graphing Software-USBDocument11 pagesInstruction of Using VSoft Viscometer Data Collection, Analysis and Graphing Software-USBBe FluentPas encore d'évaluation
- QPA TrainingDocument13 pagesQPA TrainingsenthilkumarPas encore d'évaluation
- Heap Sort Applet AnimationDocument18 pagesHeap Sort Applet AnimationSimana PeriePas encore d'évaluation
- ADS TutorialDocument7 pagesADS TutorialNithesh Chakravarthi NekkantiPas encore d'évaluation
- 01 IntroductionDocument2 pages01 IntroductionSonia GspPas encore d'évaluation
- Lab I - Introduction To Labview Objectives: Page 1 of 6Document6 pagesLab I - Introduction To Labview Objectives: Page 1 of 6Paul MusyokaPas encore d'évaluation
- Heartrate Variability AnalysisDocument23 pagesHeartrate Variability AnalysisIndia Adams100% (5)
- Compositeimageassignment Filled in For SureDocument10 pagesCompositeimageassignment Filled in For Sureapi-278321703Pas encore d'évaluation
- Image Filtering: Low and High FrequencyDocument5 pagesImage Filtering: Low and High FrequencyvijayPas encore d'évaluation
- RISA 2D Truss AnalysisDocument161 pagesRISA 2D Truss AnalysisIam engineerPas encore d'évaluation
- AUTODESK INVENTOR FEA TUTORIALDocument24 pagesAUTODESK INVENTOR FEA TUTORIALimyourscin100% (3)
- R Dap Getting StartedDocument11 pagesR Dap Getting StartedAlexPas encore d'évaluation
- MSC/PATRAN Bar Problem TutorialDocument72 pagesMSC/PATRAN Bar Problem TutorialJitendra ItankarPas encore d'évaluation
- Sap Truess Step by Step ProceduresDocument18 pagesSap Truess Step by Step ProceduresThulasirajan KrishnanPas encore d'évaluation
- 4Cast XL User Guide: Time-Series Prediction in 5 StepsDocument31 pages4Cast XL User Guide: Time-Series Prediction in 5 StepsAbhinav JainPas encore d'évaluation
- Warpalizer ManualDocument16 pagesWarpalizer ManualMrHappyHoundsPas encore d'évaluation
- WaferMapperUsersGuide1 0Document60 pagesWaferMapperUsersGuide1 0Juan Edo Rodríguez GaticaPas encore d'évaluation
- Users Guide v1Document21 pagesUsers Guide v1Joanne Ash MajdaPas encore d'évaluation
- Asfafjhaf F SDF DSFSD F Sdfs Dfsdfs FSF S Fer SBFDHD GFDGDF G D G DF GDFG DFDocument22 pagesAsfafjhaf F SDF DSFSD F Sdfs Dfsdfs FSF S Fer SBFDHD GFDGDF G D G DF GDFG DFaamirPas encore d'évaluation
- Fine Element Method Using NastranDocument84 pagesFine Element Method Using Nastranirbar.darmansyah.alwiPas encore d'évaluation
- GEO/EVS 425/525 Unit 7 Satellite Image Rectification: Step 1: Subsetting and Layerstacking Your ImageDocument6 pagesGEO/EVS 425/525 Unit 7 Satellite Image Rectification: Step 1: Subsetting and Layerstacking Your ImageDavid Andrés HerreraPas encore d'évaluation
- Actin Flow Metamorph Analysis ProcedureDocument5 pagesActin Flow Metamorph Analysis Procedurebackup.data.drive.03Pas encore d'évaluation
- TolAnalyst TutorialDocument15 pagesTolAnalyst TutorialAnton Nanchev100% (1)
- Catia V5 Bending Torsion Tension Shear TutorialDocument18 pagesCatia V5 Bending Torsion Tension Shear Tutorialrankx001Pas encore d'évaluation
- BLI ManualDocument7 pagesBLI ManualKURTPas encore d'évaluation
- FractalyseDocument13 pagesFractalyseJuan Francisco Durango GrisalesPas encore d'évaluation
- Space syntax analysis tutorialDocument10 pagesSpace syntax analysis tutorialnil julfikarPas encore d'évaluation
- M Nova Quick GuideDocument6 pagesM Nova Quick GuideALI MUHAMMADPas encore d'évaluation
- DYNAMO Walkthrough GuideDocument16 pagesDYNAMO Walkthrough GuideashPas encore d'évaluation
- Shear Strength Reduction Analysis in Phase2Document12 pagesShear Strength Reduction Analysis in Phase2Maulida Surya IrawanPas encore d'évaluation
- Flatirons Training ManualDocument38 pagesFlatirons Training Manualgsiprocessing7Pas encore d'évaluation
- Optimize species distributions with Desktop GarpDocument13 pagesOptimize species distributions with Desktop GarpTee SiPas encore d'évaluation
- GaussiantutorialDocument17 pagesGaussiantutorialkoshiPas encore d'évaluation
- Tutorial Minitab 15Document32 pagesTutorial Minitab 15wawan_a_sPas encore d'évaluation
- Micro Leveling Tech NoteDocument4 pagesMicro Leveling Tech NotemarcusnicholasPas encore d'évaluation
- Evolutionary Algorithms for Food Science and TechnologyD'EverandEvolutionary Algorithms for Food Science and TechnologyPas encore d'évaluation
- NX 9 for Beginners - Part 1 (Getting Started with NX and Sketch Techniques)D'EverandNX 9 for Beginners - Part 1 (Getting Started with NX and Sketch Techniques)Évaluation : 3.5 sur 5 étoiles3.5/5 (8)
- Oklahoma Corporation Commission: How to Download Monthly Gas Production Reports for Your Well: Landowner Internet Tutorials Series I, #2D'EverandOklahoma Corporation Commission: How to Download Monthly Gas Production Reports for Your Well: Landowner Internet Tutorials Series I, #2Pas encore d'évaluation
- Assembler, Compiler, Loader and LinkerDocument5 pagesAssembler, Compiler, Loader and LinkerBrahmanand Singh100% (1)
- Core Python by Vikash SarrafDocument153 pagesCore Python by Vikash SarrafPutluri PriyankaPas encore d'évaluation
- Marketing MCQ For Bank PODocument22 pagesMarketing MCQ For Bank POPooja Singh50% (2)
- 01 OverviewDocument65 pages01 OverviewdqltakedaPas encore d'évaluation
- User Guide: File Master 1 .1 Folder Cleaner 1 .0Document5 pagesUser Guide: File Master 1 .1 Folder Cleaner 1 .0TonyLandeoPas encore d'évaluation
- Capabilities of C++Document6 pagesCapabilities of C++npraj888312Pas encore d'évaluation
- Unit 1 Software Evolution: Structure Page NoDocument41 pagesUnit 1 Software Evolution: Structure Page NoJignesh PatelPas encore d'évaluation
- Learning Gravostyle'98: A Training ManualDocument19 pagesLearning Gravostyle'98: A Training Manualcristian cisternasPas encore d'évaluation
- 21BCS11102 - Prashant Kumar C++ 1.3Document13 pages21BCS11102 - Prashant Kumar C++ 1.3AswinPas encore d'évaluation
- Object Modeling and C++ Programming For Bca 2nd Semester PDFDocument44 pagesObject Modeling and C++ Programming For Bca 2nd Semester PDFsatyapt007Pas encore d'évaluation
- Class 10 Computer Science MCQ and Python programsDocument2 pagesClass 10 Computer Science MCQ and Python programsSobana JoyPas encore d'évaluation
- 1-Language Processing SystemDocument23 pages1-Language Processing Systemsherlock holmesPas encore d'évaluation
- OP37Document252 pagesOP37Augusto CamposPas encore d'évaluation
- Final Year Report PDFDocument56 pagesFinal Year Report PDFJay PatelPas encore d'évaluation
- Access Matrix: Implementation and ComparisonDocument19 pagesAccess Matrix: Implementation and ComparisonHamzah HamizanPas encore d'évaluation
- Accounting Information System: Midterm ExamDocument37 pagesAccounting Information System: Midterm ExamHeidi OpadaPas encore d'évaluation
- Project Report DCS 31-07-09Document111 pagesProject Report DCS 31-07-09Himanshu Verma100% (2)
- Atari 1050 ServiceDocument51 pagesAtari 1050 ServiceAdriaan StolkPas encore d'évaluation
- MSW Users Manual, TriStation v3.1Document490 pagesMSW Users Manual, TriStation v3.1guillermo muñozPas encore d'évaluation
- Lesson 2 - Introduction To JavaDocument23 pagesLesson 2 - Introduction To JavaSeth Albores Galela IIPas encore d'évaluation
- Tutorial 3 Keil DebuggerDocument6 pagesTutorial 3 Keil DebuggerNaresh BopparathiPas encore d'évaluation
- DNCDocument14 pagesDNCashoku24007Pas encore d'évaluation
- Dca1102 Programming in CDocument4 pagesDca1102 Programming in Cdaman.d778Pas encore d'évaluation
- Jain College I PUC Mock Computer Science Exam QuestionsDocument2 pagesJain College I PUC Mock Computer Science Exam QuestionsAkash RajPas encore d'évaluation
- LTpower CADhelpDocument31 pagesLTpower CADhelpMatthew TowerPas encore d'évaluation
- TE-2200-765 Dealer Manual PDFDocument357 pagesTE-2200-765 Dealer Manual PDFOscarPas encore d'évaluation
- Question Bank MCQ Questions: Shivaji University, KolhapurDocument9 pagesQuestion Bank MCQ Questions: Shivaji University, KolhapurSakshi KoliPas encore d'évaluation
- Libertad National High School: Libertad, Bunawan, Agusan Del SurDocument51 pagesLibertad National High School: Libertad, Bunawan, Agusan Del SurZaldy Osico TejadoPas encore d'évaluation
- 12th CS Practical Manual-1Document15 pages12th CS Practical Manual-1Fahad MohammedPas encore d'évaluation
- B.sc-computer-Science 2017 2018 SyllabusDocument70 pagesB.sc-computer-Science 2017 2018 SyllabusDHIVYA SPas encore d'évaluation